Making a molecular micromap: Imaging the yeast 26S proteasome at near-atomic resolution
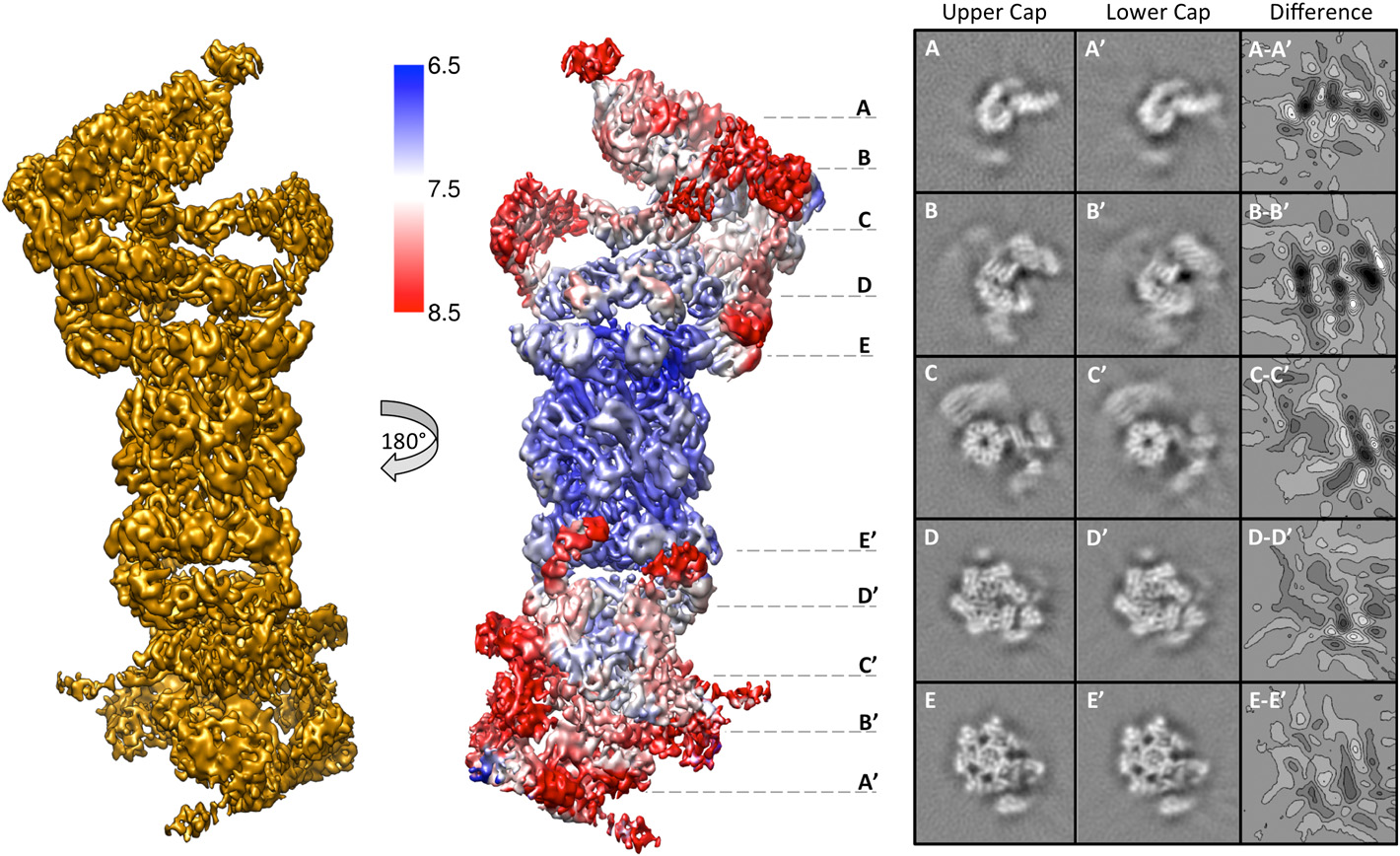

Structure of an endogenous yeast 26S proteasome reveals two major conformational states

Substrate-engaged 26S proteasome structures reveal mechanisms for ATP-hydrolysis–driven translocation

Structure of the human 26S proteasome at a resolution of 3.9 Å

Molecular and cellular dynamics of the 26S proteasome - ScienceDirect
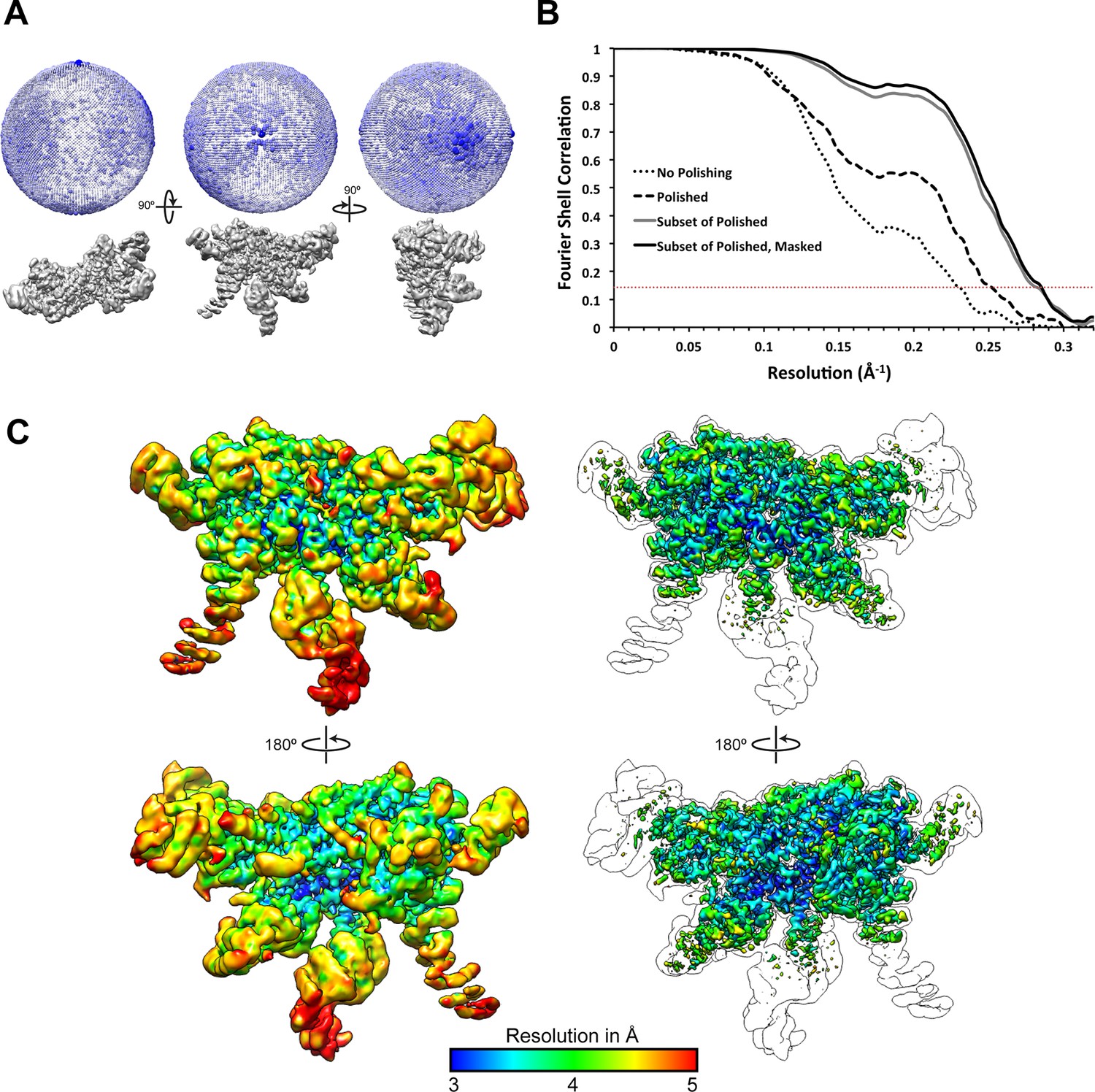
Atomic structure of the 26S proteasome lid reveals the mechanism of deubiquitinase inhibition

A molecular census of 26S proteasomes in intact neurons

Structure of an endogenous yeast 26S proteasome reveals two major conformational states

Localization of the 26S proteasome during mitosis and meiosis in fission yeast

Structure of an endogenous yeast 26S proteasome reveals two major conformational states

Molecular and cellular dynamics of the 26S proteasome - ScienceDirect

Structure of the human 26S proteasome at a resolution of 3.9 Å

Structure of an endogenous yeast 26S proteasome reveals two major conformational states
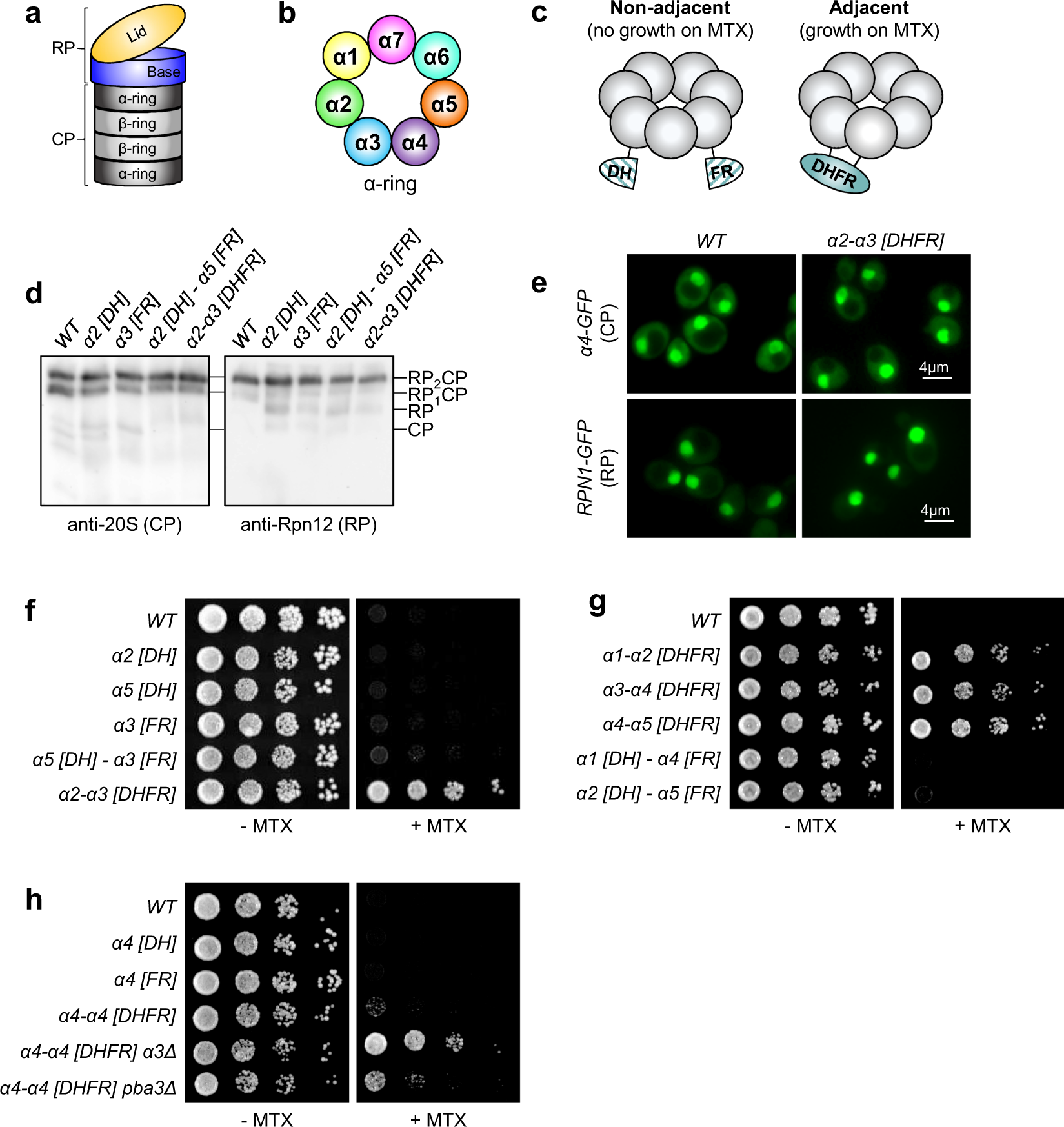
Proteasome subunit α1 overexpression preferentially drives canonical proteasome biogenesis and enhances stress tolerance in yeast

Near-atomic resolution structural model of the yeast 26S proteasome

Phosphorylation of Tyr-950 in the proteasome scaffolding protein RPN2 modulates its interaction with the ubiquitin receptor RPN13 - ScienceDirect