ChemEM: Flexible Docking of Small Molecules in Cryo-EM Structures


Cryo-EM structure of NSF in the ADP-AlFx state.(a) The 3D

Cryo-EM structure determination of the human proteasome in four
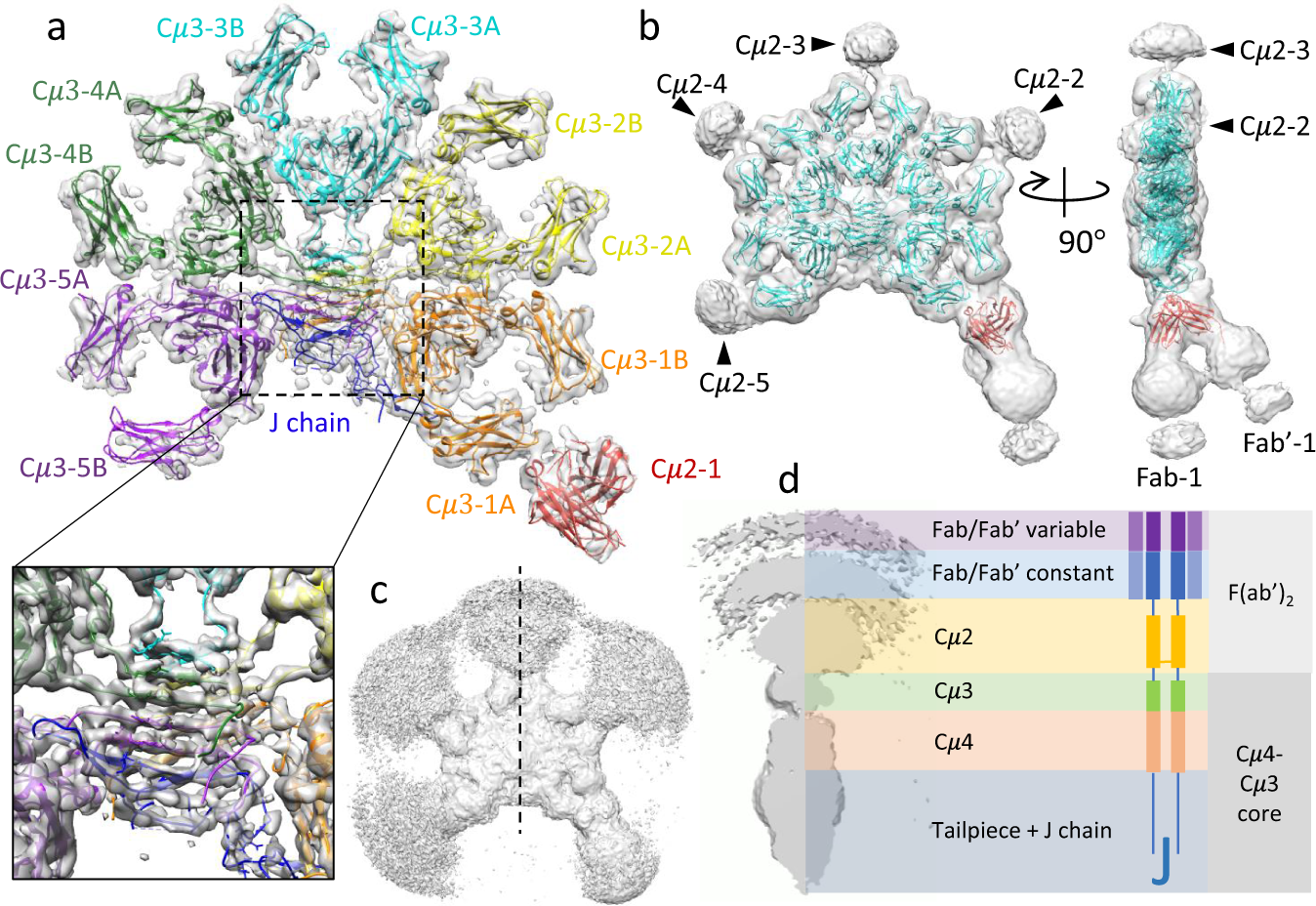
Cryomicroscopy reveals the structural basis for a flexible hinge motion in the immunoglobulin M pentamer

Cryo-EM structure of the human chemerin receptor 1–Gi protein complex bound to the C-terminal nonapeptide of chemerin

Comparing Cryo-EM Reconstructions and Validating Atomic Model Fit Using Difference Maps

ChemEM: flexible docking of small molecules in Cryo-EM structures using difference maps - Abstract - Europe PMC

Capturing protein communities by structural proteomics in a thermophilic eukaryote

ChemEM: Flexible Docking of Small Molecules in Cryo-EM Structures

Benchmarking applicability of medium‐resolution cryo‐EM protein structures for structure‐based drug design - Lee - 2023 - Journal of Computational Chemistry - Wiley Online Library

Benchmarking EMERALD against the EMDB a A comparison of EMERALD models
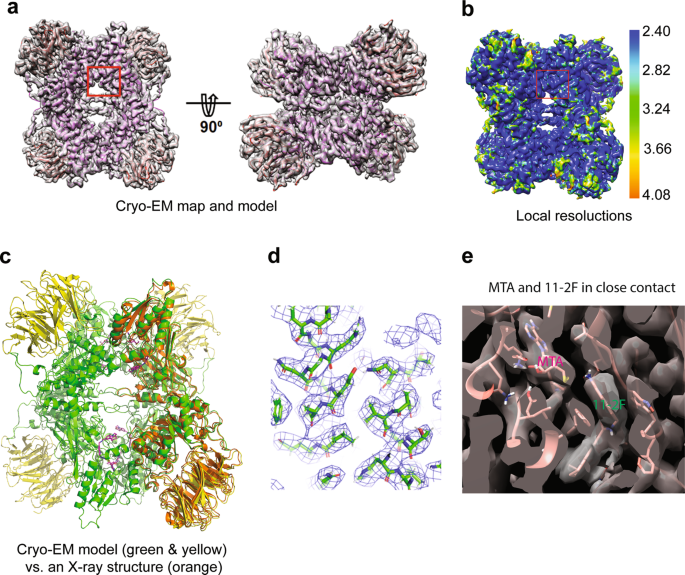
Cryo-EM structure-based selection of computed ligand poses enables design of MTA-synergic PRMT5 inhibitors of better potency

Cryo-EM structure-based selection of computed ligand poses enables design of MTA-synergic PRMT5 inhibitors of better potency
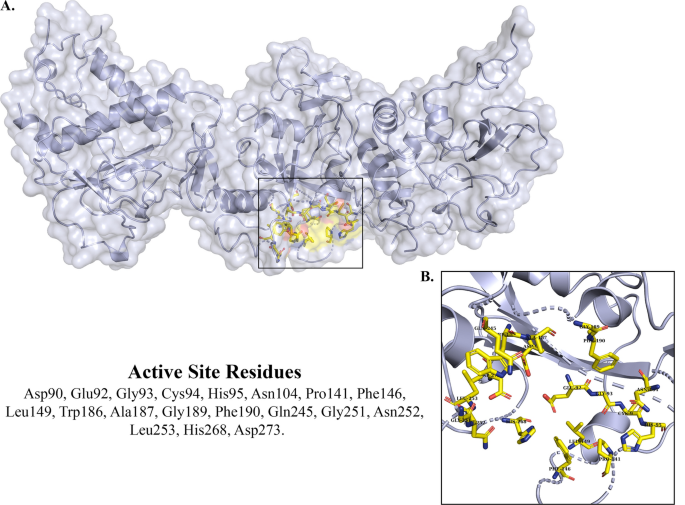
Exploring the pharmacological aspects of natural phytochemicals against SARS-CoV-2 Nsp14 through an in silico approach

Integrative Modeling of Biomolecular Complexes: HADDOCKing with Cryo-Electron Microscopy Data - ScienceDirect